B. G09 ESP Calculations with/without a Test Charge
Now prepare to perform POP optimization to determine atomic polarizability.
POP optimization requires ESP on the molecular surface and ESPs on the molecular surface when a series of test charges are placed on the molecular surface.
ESPs for molecular surfaces are created using a modified version of G09.
The calculation is performed in the following three steps.
1. Create G09 input files for ESP outputs
"makegjf_g09.cm" is a sample csh command to make gaussian job files from the rest file. The restart data of ESP ("opt_scf_C_aT.rest" in "A G09 MOD ESP" page) is required. This shell script can be run by using the command line:
"> cshrc makegjf_g09.cm"
Download
"makegjf_g09.inp" is a sample input to make gaussian job files from the rest file.
Download
"makegjf_g09.f" is a FORTRAN program to make gaussian job files from the rest file. The FORTRAN program can be compile as follows:
"> gfortran -o makegjf_g09.exe makegjf_g09.f"
The executable file is makegjf_g09.exe.
Download
After running makegjf_g09.cm, the files d0.gjf and d001.gjf-dxxx.gjf are created. d0.gjf is the G09 input file without a test charge. d001.gjf-dxxx.gjf are G09 input files with a test charge. "xxx" represents the number of unique points on the molecular surface. For Kosugi point selection, the molecular surface points for cytosine is 337. The number can be reduced to 187 using symmetry of cytosine. The position of the test charge does not necessarily have to match the position of the Kosugi point selection, but there is no problem with the same position. So the G09 input files of d001.gjf-d187.gjf are created.
"d0.gjf" is shown.
Download
"d003.gjf" is shown as an example.
Download
- Create G09 input files for ESP outputs
- Calculation of ESP with/without test charge
- Create input date for POP optimization
1. Create G09 input files for ESP outputs
"makegjf_g09.cm" is a sample csh command to make gaussian job files from the rest file. The restart data of ESP ("opt_scf_C_aT.rest" in "A G09 MOD ESP" page) is required. This shell script can be run by using the command line:
"> cshrc makegjf_g09.cm"
Download
"makegjf_g09.inp" is a sample input to make gaussian job files from the rest file.
1) 0.1 : magnitude of a test charge.
2) cc-pVTZ : basis set.
3) wB97XD : QM method. ReUse: 2 electron integral reuse key word (int=reuse).
This keyword is useful for HF calculations, but is simply ignored for DFT calculations.
4) 8 : iop(6/20=8). Kosugi 1.8 times vdw surface is selected.
5) 0 : The number of additional AOs is usually zero.
6) 0 1 : charge and multiplicity.
7) 16 : NProcShared=16. 16 cpu is selected.
2) cc-pVTZ : basis set.
3) wB97XD : QM method. ReUse: 2 electron integral reuse key word (int=reuse).
This keyword is useful for HF calculations, but is simply ignored for DFT calculations.
4) 8 : iop(6/20=8). Kosugi 1.8 times vdw surface is selected.
5) 0 : The number of additional AOs is usually zero.
6) 0 1 : charge and multiplicity.
7) 16 : NProcShared=16. 16 cpu is selected.
Download
"makegjf_g09.f" is a FORTRAN program to make gaussian job files from the rest file. The FORTRAN program can be compile as follows:
"> gfortran -o makegjf_g09.exe makegjf_g09.f"
The executable file is makegjf_g09.exe.
Download
After running makegjf_g09.cm, the files d0.gjf and d001.gjf-dxxx.gjf are created. d0.gjf is the G09 input file without a test charge. d001.gjf-dxxx.gjf are G09 input files with a test charge. "xxx" represents the number of unique points on the molecular surface. For Kosugi point selection, the molecular surface points for cytosine is 337. The number can be reduced to 187 using symmetry of cytosine. The position of the test charge does not necessarily have to match the position of the Kosugi point selection, but there is no problem with the same position. So the G09 input files of d001.gjf-d187.gjf are created.
"d0.gjf" is shown.
Download
"d003.gjf" is shown as an example.
Download
2. Calculation of ESP with/without test charge
"rung09_d0_allxx3.cm" is a sample csh command to run the Gaussian job files. Here we are only running 19 input files with a trailing 3 (dxx3.gjf). The wB97XD/cc-pVTZ calculation of cytosine takes a certain amount of time, so the amount of calculation is reduced to 1/10. It has been verified that reducing the test charge and ESP position by a factor of 10 does not significantly change the results. The procedure shown below is a little complicated, but POP optimization ("POP Fitting"), which will be described later, can greatly reduce the amount of calculation.
Download
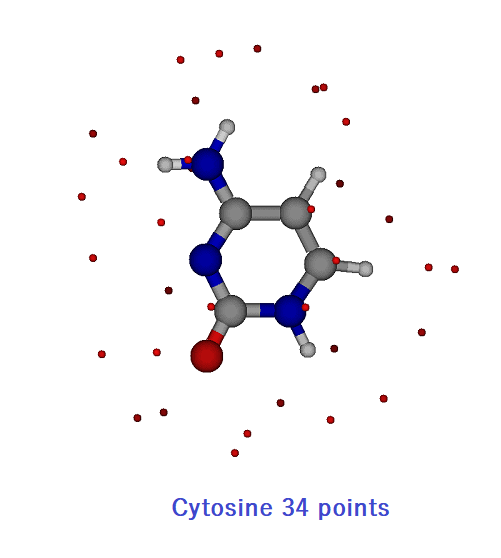
After all G09 calculations are completed, the ESP data of the molecular surface are output as rest files (d0_scf.rest and dxx3_scf.rest). Taking the differences between d0 and dxx3 give a series of polarized one-electron potentials (POPs). Here the molecular surface points of ESP are also reduced to 1/10 (337 -> 34 points). Thus, for cytosine with Cs symmetry, 19×34 points are used for POP optimization. POP optimization optimizes the atomic polarizabilities of a molecule.
Overlay 6 of G09 has an option iop(50) regarding whether to write the Antechamber file (for AMBER) during ESP charge fitting. This option doesn't seem to work when used with the "massage" keyword (No vectors left in VDWSrf). In Gaussian 16, the "massage" key word seem to be not working.
"rung09_d0_allxx3.cm" is a sample csh command to run the Gaussian job files. Here we are only running 19 input files with a trailing 3 (dxx3.gjf). The wB97XD/cc-pVTZ calculation of cytosine takes a certain amount of time, so the amount of calculation is reduced to 1/10. It has been verified that reducing the test charge and ESP position by a factor of 10 does not significantly change the results. The procedure shown below is a little complicated, but POP optimization ("POP Fitting"), which will be described later, can greatly reduce the amount of calculation.
Download
After all G09 calculations are completed, the ESP data of the molecular surface are output as rest files (d0_scf.rest and dxx3_scf.rest). Taking the differences between d0 and dxx3 give a series of polarized one-electron potentials (POPs). Here the molecular surface points of ESP are also reduced to 1/10 (337 -> 34 points). Thus, for cytosine with Cs symmetry, 19×34 points are used for POP optimization. POP optimization optimizes the atomic polarizabilities of a molecule.

Overlay 6 of G09 has an option iop(50) regarding whether to write the Antechamber file (for AMBER) during ESP charge fitting. This option doesn't seem to work when used with the "massage" keyword (No vectors left in VDWSrf). In Gaussian 16, the "massage" key word seem to be not working.
3. Create input date for POP optimization
"extredlinpot.cm" is a sample csh command. Test charge position data (extfldxyz.dat) and a series of molecular surface POP data (rest_2_3_3_C) used for POP optimization are created.
Download
"extfldxyz.dat" contains test charge position data.
Download
"rest_2_3_3_C" contains data for use in POP optimization.
Download
"extredlinpot.cm" is a sample csh command. Test charge position data (extfldxyz.dat) and a series of molecular surface POP data (rest_2_3_3_C) used for POP optimization are created.
1) Program extfldxyz.f extracts the 19 test charge position data (xx3) from the new_d0_scf.rest file and writes it to the extfldxyz.dat file.
To extract xx3, enter "3" from dat3.dat.
2) Concatenate rest files (d0_scf.rest, d003_scf.rest, d013_scf.rest ... dxx3_scf.rest). g09_scf_3.rest file is created.
3) Program extredatmpot.f deletes the ESP data to 1/10 and atoms. "red0.dat" specifies ESP data with 3 at the end and 0 atoms to be deleted.
The data used by POP optimization is stored in the rest_2_3_3_C file. The surface ESP data of 19×34 are stored.
To extract xx3, enter "3" from dat3.dat.
2) Concatenate rest files (d0_scf.rest, d003_scf.rest, d013_scf.rest ... dxx3_scf.rest). g09_scf_3.rest file is created.
3) Program extredatmpot.f deletes the ESP data to 1/10 and atoms. "red0.dat" specifies ESP data with 3 at the end and 0 atoms to be deleted.
The data used by POP optimization is stored in the rest_2_3_3_C file. The surface ESP data of 19×34 are stored.
Download
"extfldxyz.dat" contains test charge position data.
Download
"rest_2_3_3_C" contains data for use in POP optimization.
Download