









Above are photos of Japanese maples and cherry blossoms.
Photos of Japan, Sweden, Norway, and Germany are presented on the Japanese page.
Polarizable Molecular Block Model for Induced Dipole Force Field
Polarizable Molecular Block Model for Induced Dipole Force Field
When simulating the motion of highly ionized, flexible macromolecules such as DNA,
it is important to correctly evaluate the intramolecular polarization in the induced dipole force field.
The polarizable molecular block (PMB) model described here is a method for estimating the energy of macromolecules undergoing conformational changes by dividing a large molecule into several smaller molecular blocks.
The PMB model more accurately evaluates the energy of macromolecules by evaluating induced dipole-charge and induced dipole-dipole interactions between molecular blocks.
The basic idea of the PMB model is illustrated in Figure 1 using three molecules (molecule A, B, and C). Here, the small rigid molecules are represented by the rectangular building blocks. q is the electrostatic potential (ESP) optimized charge of the molecule. α is the polarized one-electron potential (POP) optimized polarizability of the molecule. These are placed on the atoms. The POPs are the differences between ESPs with and without an external test charge. When the three molecules interact, induced dipoles (μ) are generated on the atoms. Since the ESP fitted charges implicitly include intramolecularly induced dipole moments, only the dipole-charge interactions induced between molecules are evaluated, as indicated by the purple arrows in the figure. Intramolecular dipole-charge interactions are excluded. Although not shown in the figure, all induced dipole-dipole interactions have been evaluated. However, according to the definition of the induced dipole force field, a dumping function is applied to nearby dipole-dipole pairs. The induced dipole can be written as
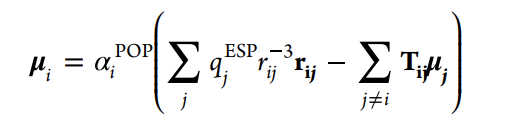
Here, qj is the charge of the molecule other than the molucule containing i. αi is the polarizability of atom i. μj is the induced dipole moment other than the induced dipole moment i. Tij is the dipole field tensor.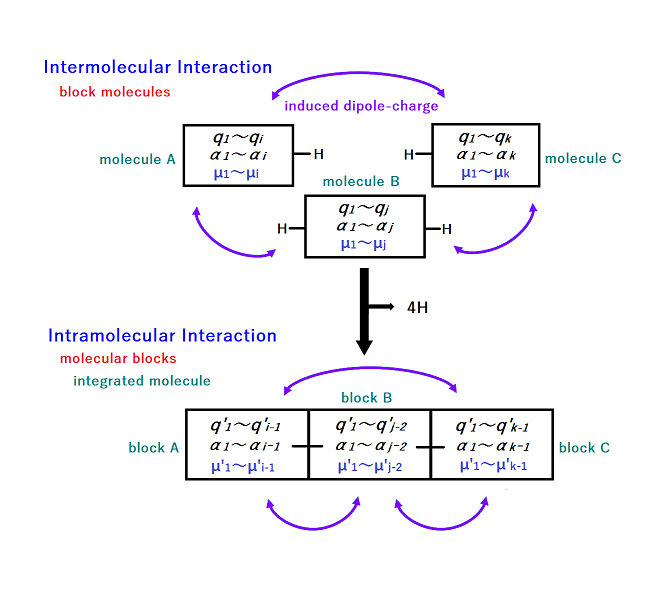
The figure shows the process of connecting three molecules together to make an integrated molecule. Four hydrogens are removed to form two single bonds. The integrated molecule consists of three molecular blocks (block A, B, and C). For integrated molecule, it seems feasible to apply the same equation. In this case, qj is the charge of the molecular blocks other than the molecular block containing i. Molecular blocks and the separated molecules are treated in the same way. However, there are some points to note in the case of the molecular blocks. The total charge of each molecular block must be strictly controlled to be an integer after the overlapping H atoms are removed. Here, the charges of the molecular block are re-optimized (q') by a capped hydrogen removal (CHR) operation. On the other hand, the atomic polarizabilities of the molecular block do not need to be adjusted, so it is used as is. The polarizabilities on the extra hydrogens are just discarded. The PMB model allows for consistent treatment of inter- and intramolecular polarization.
For more information on the PMB model, please refer to the following recently published paper.
This web page mainly describes the procedure for generating non-bonded parameters of the induced dipole force field by the polarizable molecular block (PMB) model. As an example, the parameters of the minimal model of DNA, the trinucleotide, are determined.
The basic idea of the PMB model is illustrated in Figure 1 using three molecules (molecule A, B, and C). Here, the small rigid molecules are represented by the rectangular building blocks. q is the electrostatic potential (ESP) optimized charge of the molecule. α is the polarized one-electron potential (POP) optimized polarizability of the molecule. These are placed on the atoms. The POPs are the differences between ESPs with and without an external test charge. When the three molecules interact, induced dipoles (μ) are generated on the atoms. Since the ESP fitted charges implicitly include intramolecularly induced dipole moments, only the dipole-charge interactions induced between molecules are evaluated, as indicated by the purple arrows in the figure. Intramolecular dipole-charge interactions are excluded. Although not shown in the figure, all induced dipole-dipole interactions have been evaluated. However, according to the definition of the induced dipole force field, a dumping function is applied to nearby dipole-dipole pairs. The induced dipole can be written as
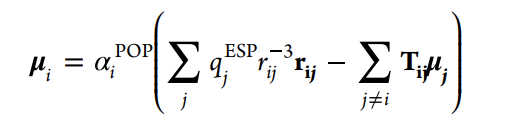
Here, qj is the charge of the molecule other than the molucule containing i. αi is the polarizability of atom i. μj is the induced dipole moment other than the induced dipole moment i. Tij is the dipole field tensor.

Figure 1. Consistent treatment of inter- and intramolecular polarization in the PMB model.
q is the ESP-optimized charge of the molecule. q' is the ESP-optimized charge of the molecular block. Four hydrogens are removed to form two single bonds.
α is the POP-optimized polarizability of the the molecule.
μ and μ' are the induced dipoles of the block molecule and the molecular block, respectively.
The figure shows the process of connecting three molecules together to make an integrated molecule. Four hydrogens are removed to form two single bonds. The integrated molecule consists of three molecular blocks (block A, B, and C). For integrated molecule, it seems feasible to apply the same equation. In this case, qj is the charge of the molecular blocks other than the molecular block containing i. Molecular blocks and the separated molecules are treated in the same way. However, there are some points to note in the case of the molecular blocks. The total charge of each molecular block must be strictly controlled to be an integer after the overlapping H atoms are removed. Here, the charges of the molecular block are re-optimized (q') by a capped hydrogen removal (CHR) operation. On the other hand, the atomic polarizabilities of the molecular block do not need to be adjusted, so it is used as is. The polarizabilities on the extra hydrogens are just discarded. The PMB model allows for consistent treatment of inter- and intramolecular polarization.
For more information on the PMB model, please refer to the following recently published paper.
S. Nakagawa, A. Kimura, Y. Okamoto, Polarizable Molecular Block Model: Toward the Development of an Induced Dipole Force Field for DNA.
J. Phys. Chem. B, 2022, 126, 10646-10661.
J. Phys. Chem. B, 2022, 126, 10646-10661.
This web page mainly describes the procedure for generating non-bonded parameters of the induced dipole force field by the polarizable molecular block (PMB) model. As an example, the parameters of the minimal model of DNA, the trinucleotide, are determined.
- G09 Mod ESP: Make Surface ESP by G09
- G09 Tst Chg: G09 Calculation of ESP with/without a Test Charge
- ESP Fitting: Electrostatic Potential Fitting
- POP Fitting: Polarized One-electron Potential Fitting
- IDFF PMB: Computation of Polarizable Molecular Block Model Using Non-bonded Induced Dipole Force Field
- IDFF TopPar: Creation of Topology & Parameter File for Polarizable Molecular Block Model
My Current Situation
From April 2022, I have been conducting joint research with researchers at the Graduate School of Science, Nagoya University. We are currently submitting a paper to foreign journals. I often do my research at home.
The submitted paper was accepted.(S. Nakagawa, A. Kimura, Y. Okamoto, Polarizable Molecular Block Model: Toward the Development of an Induced Dipole Force Field for DNA. J. Phys. Chem. B, 126, 10646-10661 (2022))
From April 2022, I have been conducting joint research with researchers at the Graduate School of Science, Nagoya University. We are currently submitting a paper to foreign journals. I often do my research at home.
The submitted paper was accepted.(S. Nakagawa, A. Kimura, Y. Okamoto, Polarizable Molecular Block Model: Toward the Development of an Induced Dipole Force Field for DNA. J. Phys. Chem. B, 126, 10646-10661 (2022))
From April 2021, I conducted research as a visiting researcher at the Graduate School of Science, Nagoya University.
The theme is "Development of DNA Induced dipole force field based on polarizable molecular block model".
In December 2020, I became Professor Emeritus of Kinjo Gakuin University.
For one year from April 2020, I was a visiting professor at the Graduate School of Science, Nagoya University.
I gave a series of lectures on zoom due to SARS-CoV-2.
(content) 1) Structure and Function of Nucleic Acid (introduction)
2) Structure and Function of Cell / Cell Membrane
3) Structure and Function of Cell / Intracellular Organelles 1
4) Structure and Function of Cell / Intracellular Organelles 2
5) Structure and Function of Cell / Cytoskeleton & Cell Junctions
6) Development of Polarizable Force Field for Nucleic Acids 1 / Quantum Chemical studies
7) Development of Polarizable Force Field for Nucleic Acids 2 / Towards a Polarizable Building Block Model
8) Structure and Function of Nucleic Acid / DNA and Genome editing
9) Structure and Function of Nucleic Acid / Genomes, Chromosomes,
DNA Replication
10) Structure and Function of Nucleic Acid / Transcription, Translation and mRNA Vaccine
11) Development of Polarizable Force Field for Nucleic Acids 3 / Polarizable Building Block Model
At the end of March 2020, I will retire from Kinjo Gakuin University, where I worked for 24 years.
The final lecture will be held at 15:30 on Thursday, January 30, 2020 in Room W2-106.
A tea party will be held after the final lecture. I posted the poster below.
Seminar graduates can participate in a tea party. Please join us after inviting everyone.
I'm looking forward to seeing you. (2020.1.16)


From April 2013 to September 2013, I was a visiting professor at Heinrich-Heine-University (HHU) in Germany.
The photos shown in Japaneae Page were taken in Germany at the time.
From April 2006 to March 2007, I was a visiting professor at the Royal Institute of Technology (KTH) in Sweden.
The photos shown in Japaneae Page were taken in Sweden and Norway at the time.